Note
Go to the end to download the full example code.
Importance Sampling Cook-Torrance#
import numpy as np
import pyvista as pv
import mirage as mr
import mirage.vis as mrv
N = mr.hat(np.array([0.0, 0.0, 1.0]))
# n = mr.rand_unit_vectors(1)
O = mr.hat(np.array([0.0, -1.0, 1]))
n_samples = int(1e5)
Os = np.tile(O, (n_samples, 1))
Ns = np.tile(N, (n_samples, 1))
Ls = mr.rand_cone_vectors(N, np.pi / 2, n_samples)
brdf = mr.Brdf('cook-torrance', cd=0.0, cs=1.0, n=0.05)
a2 = brdf.n**2
frs = brdf.eval(Ls, Os, Ns)
integrand = frs * mr.dot(Ns, Ls)
print(np.sum(integrand) / n_samples * 2 * np.pi)
pl = pv.Plotter()
pl.add_mesh(pv.Plane(direction=N))
mrv.plot_arrow(pl, (0, 0, 0), N)
mrv.plot_arrow(pl, O, -O, color='green')
mrv.scatter3(pl, 0.5 * Ls, scalars=frs, point_size=10, opacity=0.2)
pl.show()
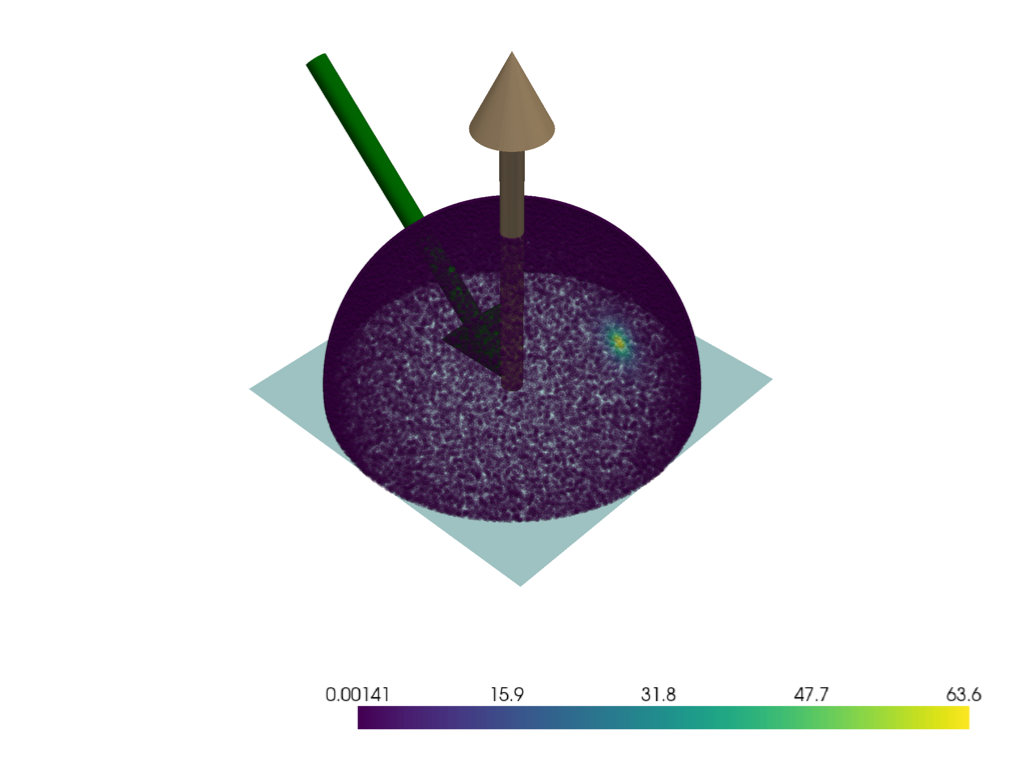
0.9698925975655143
Now that we have a good estimate of the true integral value, let’s begin down the road to importance sampling by understanding the Beckmann normal distribution function (NDF).
e1 = np.random.rand(n_samples)
e2 = np.random.rand(n_samples)
# theta = np.arccos(np.sqrt((1-e1)/(e1*(a2-1)+1))) # GGX
theta = np.arctan(np.sqrt(a2 * e1 / (1 - e1))) # GGX
phi = 2 * np.pi * e2
n_tangent = np.vstack(mr.sph_to_cart(phi, np.pi / 2 - theta)).T
x = mr.hat(np.cross(mr.rand_unit_vectors(1), N))
y = np.cross(N, x)
dcm = np.vstack((x, y, N)).T
wm = mr.stack_mat_mult_vec(dcm, n_tangent) # The micro normal vector
wo = Os
wi = mr.reflect(wo, wm)
F = mr.cook_torrance_fresnel_schlick(wm, wi, brdf.cs)
G2 = mr.cook_torrance_g_smith(wi, Ns, wo, np.sqrt(a2))
weight = mr.dot(wo, wm) / (mr.dot(wo, Ns) * mr.dot(wm, Ns))
integrand_importance = F * G2 * weight
ok = (mr.dot(Ns, wi) > 0.0) & (mr.dot(wi, wm) > 0.0)
n_used_samples = np.sum(ok)
print(np.sum(integrand_importance[ok]) / n_used_samples)
# brdf_vals = brdf.eval(wis, wo_, ns).flatten()
# brdf_vals[is_below_horizon] = 0
# print(np.sum(brdf_vals/p)/n_samples)
# endd
pl = pv.Plotter()
pl.add_mesh(pv.Plane(direction=N))
mrv.plot_arrow(pl, (0, 0, 0), N)
mrv.plot_arrow(pl, O, -O, color='green')
# mrv.scatter3(pl, 0.5*wos, scalars=ds, point_size=10)
mrv.scatter3(pl, wi[ok.flatten(), :], point_size=5, opacity=0.2, scalars=weight[ok])
mrv.scatter3(pl, wm, point_size=10, opacity=0.1, scalars=ok, cmap='plasma')
pl.show()
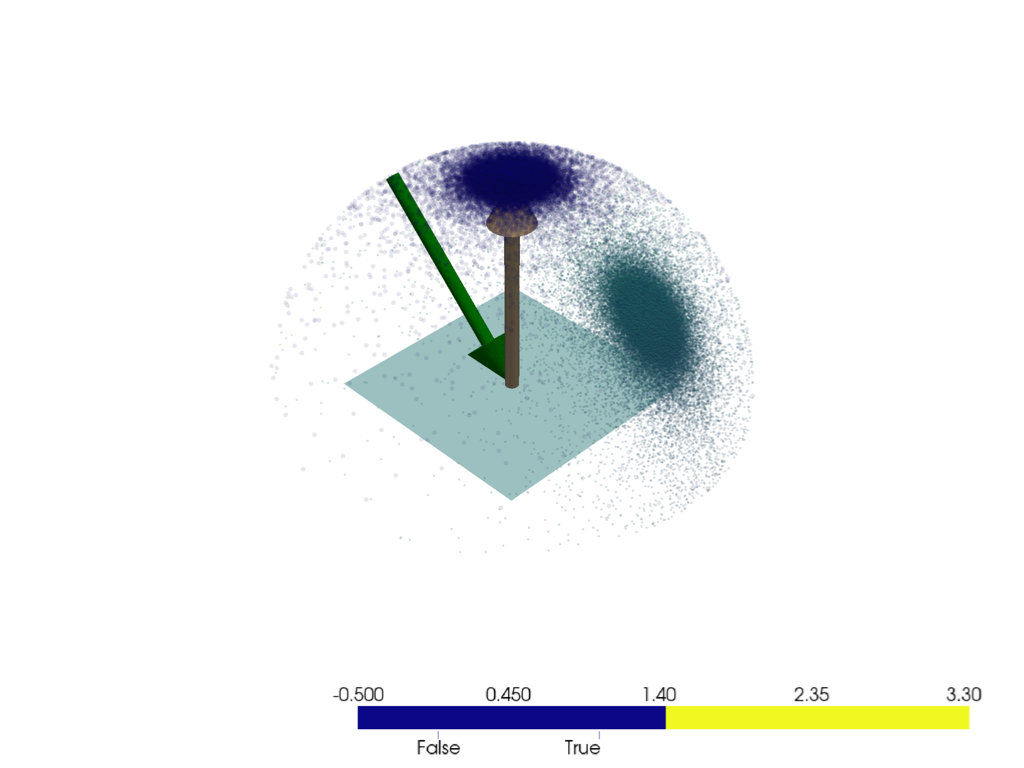
1.0006715797881398
Total running time of the script: (0 minutes 0.398 seconds)
